
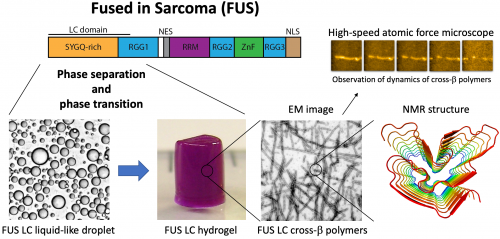
 Intercellular organelles, such as nuclei, mitochondria, endoplasmic reticulum and Golgi, have lipid bi-layer membranes, which compartmentalize the inner space of these organelles from outside environments to facilitate their cellular functions. Other than these organelles, cells also have sub-cellular structures that are not surrounded by membranes. For example, RNA granules are condensates formed from RNA and many RNA-binding proteins. RNA granules play important cellular functions in RNA metabolisms such as storage, degradation or transportation. For a long time, little had been known about the mechanisms for the formation of non-membrane bound structures. Recently, it was shown that liquid-liquid phase separation (LLPS: a physical phenomenon such as an oil phase separates from a water phase) of proteins and RNAs could be an underlying mechanism. However, molecular details during a phase transition of proteins/RNAs to the phase separated state are largely unknown. Proteins/RNAs in the phase separated state are highly dynamic. Therefore, a study of molecular details is difficult with conventional biophysical techniques. We will use the state-of-the-art quantum techniques such as nano-diamond sensors, neutron scattering and high-speed atomic force microscope, which can observe the real-time movement of molecules to understand the molecular mechanisms of LLPS. It is known that mutations of some of the RNA-binding proteins, such as FUS and TDP-43, that accumulate in RNA granules are causative of neurodegenerative diseases including amyotrophic lateral sclerosis and frontotemporal degeneration. It is hypothesized that disease mutations of these proteins cause an aberrant phase separation, leading to dysfunction of RNA granules and formation of fibrous aggregates and thus neurodegenerative diseases. We aim to contribute to developing treatment methods for neurodegenerative diseases by studying a molecular basis causing an aberrant intracellular phase separation.
Intercellular organelles, such as nuclei, mitochondria, endoplasmic reticulum and Golgi, have lipid bi-layer membranes, which compartmentalize the inner space of these organelles from outside environments to facilitate their cellular functions. Other than these organelles, cells also have sub-cellular structures that are not surrounded by membranes. For example, RNA granules are condensates formed from RNA and many RNA-binding proteins. RNA granules play important cellular functions in RNA metabolisms such as storage, degradation or transportation. For a long time, little had been known about the mechanisms for the formation of non-membrane bound structures. Recently, it was shown that liquid-liquid phase separation (LLPS: a physical phenomenon such as an oil phase separates from a water phase) of proteins and RNAs could be an underlying mechanism. However, molecular details during a phase transition of proteins/RNAs to the phase separated state are largely unknown. Proteins/RNAs in the phase separated state are highly dynamic. Therefore, a study of molecular details is difficult with conventional biophysical techniques. We will use the state-of-the-art quantum techniques such as nano-diamond sensors, neutron scattering and high-speed atomic force microscope, which can observe the real-time movement of molecules to understand the molecular mechanisms of LLPS. It is known that mutations of some of the RNA-binding proteins, such as FUS and TDP-43, that accumulate in RNA granules are causative of neurodegenerative diseases including amyotrophic lateral sclerosis and frontotemporal degeneration. It is hypothesized that disease mutations of these proteins cause an aberrant phase separation, leading to dysfunction of RNA granules and formation of fibrous aggregates and thus neurodegenerative diseases. We aim to contribute to developing treatment methods for neurodegenerative diseases by studying a molecular basis causing an aberrant intracellular phase separation.

Team Leader, Masato Kato
Members
Researcher, Nobuo Maita
Researcher, Kim Sunyong
Technician, Yuko Kajino
Recent Publications
Fonda, D.B., Kato, M., Li, Y. and Murray, D.T. Cryo-EM and solid state NMR together provide a more comprehensive structural investigation of protein fibrils. Protein Sci. Oct;33(10):e5168, doi: 10.1002/pro.5168 (2024)
Fujii, K., Izumi, Y., Maita, N., Matsuo, K., Kato, M. Observation of the Liquid-Liquid Phase Separation of FUS-LC Using Vacuum-Ultraviolet Circular Dichroism Spectroscopy. Chirality, Aug;36(8):e23707, doi: 10.1002/chir.23707 (2024)
Kato, M. 「量子センサーと分光学を用いた相分離液滴の構造ダイナミクス研究」, 医学のあゆみ, Vol.290, No. 4, 25078-83 (2024)
Gu, J., Zhou, X., Sutherland, L., Kato, M., Jaczynska, K., Rizo, J. and McKnight, S.L. Oxidative regulation of TDP-43 self-association by a β-to-α conformational switch. Proc. Nat. Acad. Sci. 120 (41) e2311416120; https://doi.org/10.1073/pnas.2311416120 (2023)
Kato, M., “Molecular Mechanisms Defining the Structural Basis for Self-Association of the FUS Low-Complexity Domain”, Riki Kurokawa, ed., “Phase Separation in Living Cells”, Springer, Singapore, https://doi.org/10.1007 (2023)
Zhou, X., Kato, M., and McKnight, S.L. How do disordered head domains assist in the assembly of intermediate filaments? Curr. Opin. Cell Biol. 85, https://doi.org/10.1016/j.ceb.2023.102262 (2023)
加藤昌人 「神経変性疾患変異による線維凝集体形成の構造学的機構 ―low-complexityドメインの機能に与える影響」 小野賢二郎/編 「いま新薬で加速する神経変性疾患研究 異常タンパク質の構造、凝集のしくみから根本治療の真の標的に迫る」 実験医学増刊 Vol.41 No.12 ISBN 978-4-7581-0412-8 (2023)
Zhou, X., Sumrow, L., Tashiro, K., Sutherland, L., Liu, D., Qin, T., Kato, M., Liszczak, G., McKnight, S.L. Mutations linked to neurological disease enhance self-association of low-complexity protein sequences. Science, Vol 377, Issue 6601, DOI: 10.1126/science.abn5582 (2022)
加藤昌人,白木賢太郎,中川真一/編 「フロントランナー直伝 相分離解析プロトコール」 実験医学別冊 最強のステップUPシリーズ ISBN 978-4-7581-2259-7 (2022)
Kato, M., Zhou, X., and McKnight, S.L. How do protein domains of low sequence complexity work? RNA. 28: 3-15, doi: 10.1261/rna.078990.121 (2022)
Kato, M.*, and McKnight, S.L. The low-complexity domain of the FUS RNA binding protein self-assembles via the mutually exclusive use of two distinct cross-β cores. Proc. Nat. Acad. Sci. 118, e2114412118; https://doi.org/10.1073/pnas.2114412118 (2021) (* corresponding authors)
廣瀬哲郎,加藤昌人,中川真一/編 「相分離 メカニズムと疾患」 実験医学増刊 Vol.39 No.10 ISBN 978-4-7581-0395-4 (2021)
Kato, M., Tu, B.P. and McKnight, S.L. Redox-mediated regulation of low complexity domain self-association. Curr Opin Genet Dev. (2021) Jan 14;67:111-118. doi: 10.1016/j.gde.2020.12.006.
