Outline of Research
Our body is made up of 60 trillion cells, which are replaced periodically (3-4 months) by metabolism. This metabolism is based on the genetic information in DNA, the "blueprint of life" in the cell nucleus. This DNA is damaged by various factors and is repeatedly repaired. Understanding of DNA damage and its repair would lead to a better treatment and a more effective prevention of "cancer," which is derived from an abnormal cell growth.
1. Analysis of DNA damage induced by lasers and laser-driven quantum beams
We are investigating how DNA damage, which is considered to be the cause of biological effects, is produced and the mechanism of its generation by irradiating lasers and even laser-driven quantum beams.
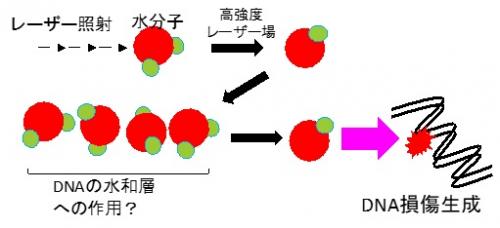
Figure 1. Analysis of laser-induced DNA damage generation
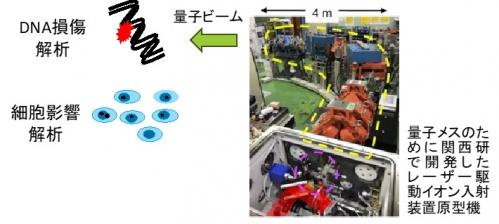
Figure 2. DNA damage generation by laser-driven quantum beam and analysis of biological effect
2. Analysis of DNA damage repair
With advanced technology utilizing fluorescence resonance energy transfer or atomic force microscopy, we are conducting research to clarify the process of DNA damage formation, its form and structure, as well as the repair process. In addition, we are also working to elucidate the mechanisms of how DNA damage leads to mutagenesis. The results obtained from these studies are expected to be useful in the treatment and prevention of cancer.
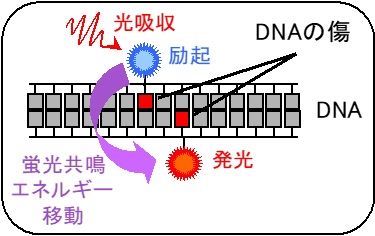
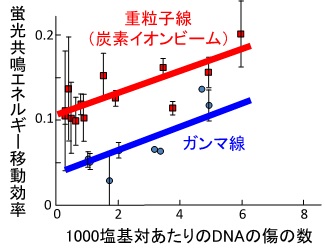
Figure 3 Localization of DNA damage measured by fluorescence resonance energy transfer
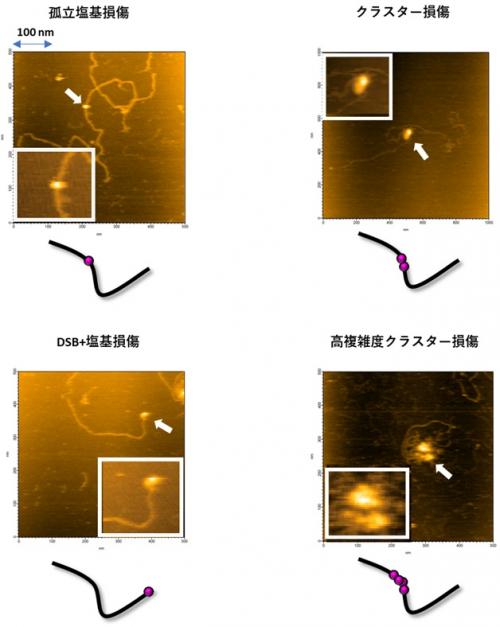
Figure 4 Various forms of DNA damage visualized using atomic force microscopy
Member
| SHIKAZONO Naoya | Project Leader |
| AKAMATSU Ken | Senior Principal Researcher |
| NAKANO Toshiaki | Senior Principal Researcher |
| MORIBAYASHI Kengo | Senior Staff |
| MASUMOTO Kotaro | QST Research Assistant |
| AOKI Hiroyo | Assistant |
| TSUDA Kanae | Assistant |
List of Achievements (Papers, Awards, Press Releases, etc.)
2023
- Yusuke Sato, Yoshihide Takaku, Toshiaki Nakano, Ken Akamatsu, Dai Inamura, and Seiichi Nishizawa, Synthetic DNA binders for fluorescence sensing of thymine glycol-containing DNA duplexes and inhibition of endonuclease activity, Chemical Communications, 59(40), 6088-6091
- Yusuke Matsuya, Yuji Yoshii, Tamon Kusumoto, Ken Akamatsu, Yuho Hirata, Tatsuhiko Sato, Takeshi Kai, A step-by-step simulation code for estimating yields of water radiolysis species based on electron track-structure mode in the PHITS code, Physics in Medicine & Biology, 69(3), ad199b
- Ken Akamatsu, Katsuya Satoh, Naoya Shikazono, and Takeshi Saito, Proximity estimation and quantification of ionizing radiation-induced DNA lesions in aqueous media using fluorescence spectroscopy, Radiation Research, 201(2), 150-159
- Kengo Moribayashi, Hiroki Matsubara, Yoshiteru Yonetani, Naoya Shikazono, Multi-Scale Simulations Aiming To Advance Heavy Ion Beam Cancer Therapy, API conference proceedings, 2756(1), 030001/1-8
- Toshiaki Nakano, Ken Akamatsu, Naoya Shikazono, Structural analysis and repair kinetics of radiation-induced DNA damage, Radiation Chemistry


